There is no doubt that molecular biologic technologies such as PCR testing have dramatically increased the sensitivity of diagnostic tests designed to detect the presence of several infectious pathogens including Streptococcus equi subsp. equi (Strep equi), Salmonella sp., CEMO and Dermatophytosis. Following on from an outbreak of strangles it is important to establish freedom from infection in all affected animals before “a line is drawn” under the outbreak and horses are free to mix and attend shows. Evidence suggests that guttural pouches are the only logical target for post-outbreak sampling with latest research indicating a greater than 50 times chance of detecting the presence of Strep equi in the guttural pouches of carriers compared to their nasopharynx (Boyle and others 2017). Furthermore, recent studies indicate that around 40% of culture-negative strangles submissions may be positive by PCR (Boyle and others 2017; Pusterla and others 2018).
However, the main drawback to reliance on PCR testing is that the technique simply detects the presence or absence of the DNA or RNA of the organism in question, whether or not that material is present in live or dead organisms. Indeed one recent study indicated that less than 20% of PCR positive and culture negative samples contained viable organisms (Pusterla and others 2018). It has been suggested that the lack of mucociliary clearance mechanisms in the guttural pouch could facilitate retention of DNA from dead Strep equi organisms leading to positive PCR results that do not actually indicate any risk of contagion.
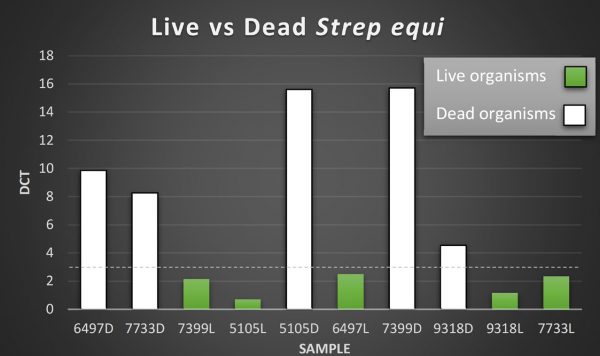
A new PCR assay has been developed at Liphook which can distinguish live from dead Strep equi organisms. Use of this PCR will enable much greater confidence in the clinical relevance of positive PCR results. Results from examination of 10 washes, 5 of which had been boiled to kill the organisms, is shown in the graph below. The PCR was able to distinguish the live from dead organisms. Further validation in greater numbers of samples is ongoing.
In order to avoid the possible confounding effect of death of organisms between sampling and testing, it is important that samples are shipped in chilled saline (ACTH chiller packs for example), as previous evidence indicates that viability of Strep equi is excellent in cold and wet conditions (Durham and others 2018).
BOYLE, A. G., STEFANOVSKI, D. & RANKIN, S. C. (2017) Comparison of nasopharyngeal and guttural pouch specimens to determine the optimal sampling site to detect Streptococcus equi subsp equi carriers by DNA amplification. BMC Vet Res 13, 75
DURHAM, A. E., HALL, Y. S., KULP, L. & UNDERWOOD, C. (2018) A study of the environmental survival of Streptococcus equi subspecies equi. Equine Vet J
PUSTERLA, N., LEUTENEGGER, C. M., BARNUM, S. M. & BYRNE, B. A. (2018) Use of quantitative real-time PCR to determine viability of Streptococcus equi subspecies equi in respiratory secretions from horses with strangles. Equine Vet J 50, 697-700